Will There be a Raftable Release out of McPhee Reservoir
I live next to the Dolores River. It’s an often overlooked gem of the southwest. It runs from just outside Rico, Colorado at its headwaters to the Colorado River near Moab, Utah. Rafting it is an experience.
As of late the river has become something of a destination for the rafting community. Most sections of the river are mellow and scenic. But two sections have class 4 rapids that keep things interesting: Snaggletooth Rapid and State Line Rapid (also known as the Chicken Raper Rapid).
In 1985 the McPhee Dam was completed, forever changing the flow of the lower Dolores River. Today, farmers, ranchers adn local ranchers rely on water from the McPhee Dam to to grow crops and raise livestock. As a result, the primary goal of reservoir managers is to keep as much water in the reservoir as possible. Raftable and ecological releases are secondary.This has resulted in several years where the reservoir has not filled and there have been no raftable releases. Other years reservoir managers say that the there will be no release and we get a release.
Because the Dolores is so loved in our community, there are a few people who try and predict when the Dolores will have a raftable release. Given my love of R, I thought it would be fun to do myself. The following are notes on how I built my model. I do my best to go over each step and detail why and how I included each covariate.
Load Packages
Here I load packages and set a custom theme. You can skip this part and just add in set_theme(theme_light) if you want to skip this part.
library(tidyverse) ## For all the data cleaning work.
library(lubridate) ## For working with dates.
library(extrafont) ## For Font Graphics
options(scipen = 999)
t<-theme_light()+
theme(
text=element_text(family = "Poppins", color = "#FFFFFF"),
plot.margin = unit(c(0.5,0.5,0.5,0.5), "cm"),
plot.title = element_text(face = "bold", size = rel(2.3), color = "#FFFFFF"),
plot.subtitle = element_text(color = "#7A7A7A", size = rel(1.3), margin = margin(t = 0, r = 0, b = 15, l = 0)),
plot.background = element_rect(fill = "#222222", color = "#FFFFFF"),
panel.grid.major = element_line(color = "#555555"),
panel.grid.minor = element_blank(),
panel.background = element_rect(fill = "#222222", color = "#222222"),
panel.border = element_blank(),
axis.text = element_text(size = rel(0.8), colour = "#CCCCCC"),
axis.title.x = element_text(margin = margin(t = 5, r = 0, b = 10, l = 0)),
axis.title.y = element_text(margin = margin(t = 0, r = 10, b = 0, l = 20)),
legend.title = element_text(face = "bold"),
legend.text = element_text(color = "#222222"),
legend.direction = "horizontal",
legend.position="bottom",
legend.key.width = unit(5, "cm"),
legend.key.height = unit(0.3, "cm"),
legend.box = "vertical",
plot.caption = element_text(color = "#7A7A7A", size = rel(0.9))
)
theme_set(t)Getting a variable to predict
Getting Stream Guage Data Below McPhee Dam from the USGS
url<-paste0("https://waterservices.usgs.gov/nwis/dv/?format=rdb&sites=09169500,%2009166500&startDT=1985-02-01&endDT=", Sys.Date(), "&statCd=00003&siteType=ST&siteStatus=all")
flow_data<-read_tsv(url, skip = 35)%>%
select(2:5)%>%
rename(site_id = 1, date = 2, flow=3, code = 4)%>%
mutate(site_id = ifelse(site_id == "09166500", "Dolores", "Bedrock"))%>%
drop_na()
bedrock_flow<-flow_data%>%
filter(site_id == "Bedrock"& year(date)>1986)%>%
select(-code)
bedrock_flow## # A tibble: 12,098 x 3
## site_id date flow
## <chr> <date> <dbl>
## 1 Bedrock 1987-01-01 120
## 2 Bedrock 1987-01-02 130
## 3 Bedrock 1987-01-03 120
## 4 Bedrock 1987-01-04 120
## 5 Bedrock 1987-01-05 140
## 6 Bedrock 1987-01-06 140
## 7 Bedrock 1987-01-07 130
## 8 Bedrock 1987-01-08 130
## 9 Bedrock 1987-01-09 110
## 10 Bedrock 1987-01-10 95
## # … with 12,088 more rowsI want to predict the raftable days released below McPhee Dam. To do this I need flow data from a gauge below McPhee. Flow gauge data will tell me which days had enough flow to raft and which days did not. There are a few gauges to to choose from within the extensive USGS flow gauge system, but the oldest is at Bedrock, CO. To get the data I used the USGS Water Services REST API. There’s an R package that you can used to get the data, but since I’d used the API a few times before and I knew that the data could be returned in tab-separated format, I just used the readr::read_tsv() function.
Walk Through Notes:
- The first step is to build the URL for the API. I used the USGS Rest API builder tool to get the right data.
- Then used the
read_tsv()function to pull data from the API skpping the first 35 lines of comments. - I then renamed the columns to something I could understand.
- And then convert the site_ids to Characters instead of numbers. I pulled data from above and below the dams because at the time I thought I might use the data from above the Dam later for another predictive variable.
- Then I drop all the missing values and subset the data to just below the dam.
Visualizing the flow
bedrock_flow%>%
filter(year(date)>2007)%>%
ggplot(aes(date, flow))+
geom_line(color = "#1AB8FF", size =1.2, alpha = 0.6)+
labs(title = "Dolores Flows from 2008 to Present",
subtitle = "USGS Flow Gauge Site 09169500 Near Bedrock, Colorado",
caption = "Data aquired from the Bureau of Reclemation using the RNRCS package.",
y = "Flow Rate (cfs)",
x = "Year")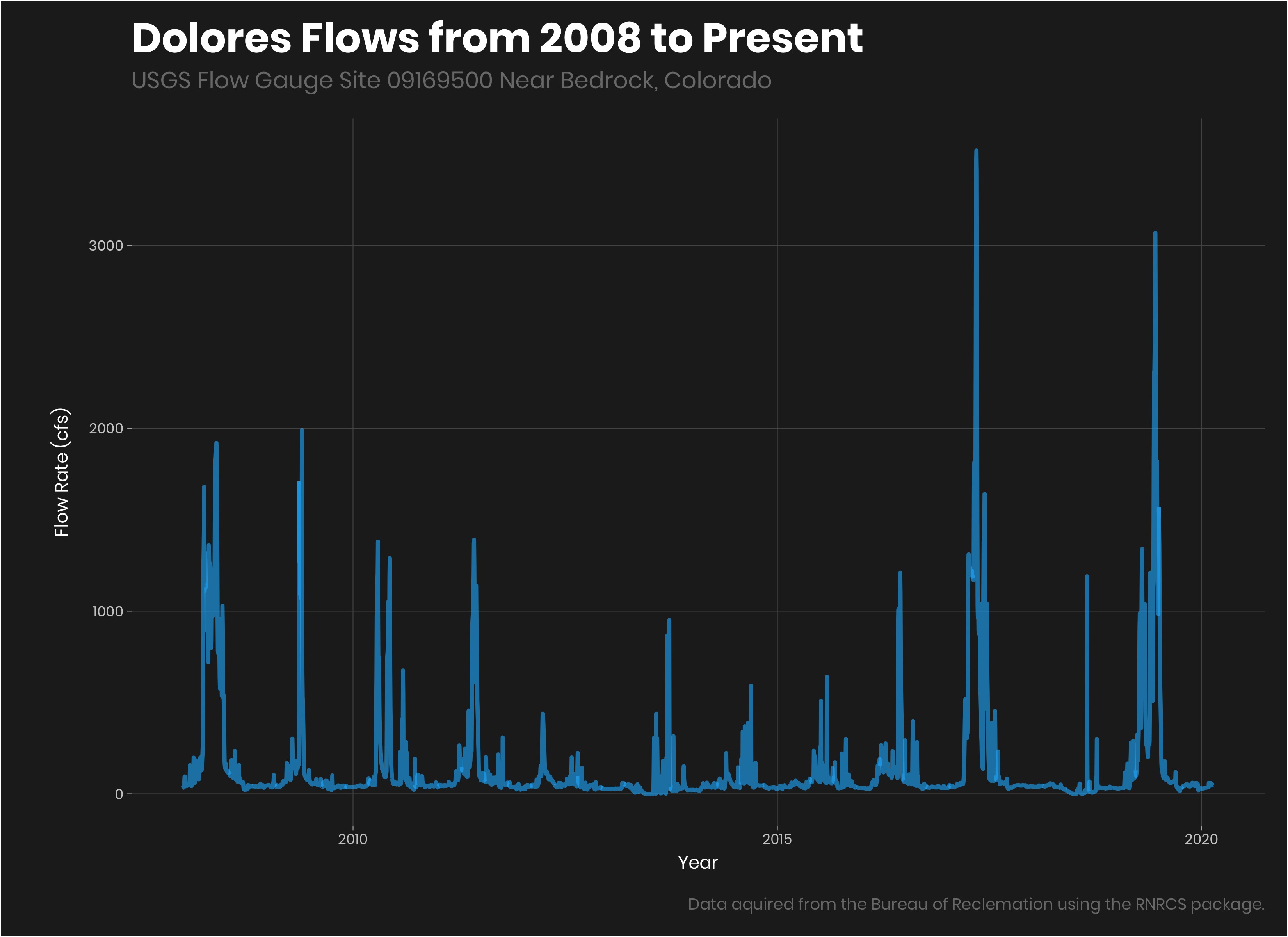
Subsetting the release
Next I needed to subset days where there is a raftable release. Releases happen during spring runoff so I wanted to subset those days in the spring that had greater than 800 cfs (cubic feet per second) of flow. I wanted to make sure to exclude non
predicted_variable<-bedrock_flow%>%
filter(flow>800 & month(date) %in% c(3:7))%>%
count(year(date))%>%
rename(year = 1, raftable_releases = 2)
predicted_variable## # A tibble: 21 x 2
## year raftable_releases
## <dbl> <int>
## 1 1987 87
## 2 1988 13
## 3 1989 17
## 4 1991 9
## 5 1992 61
## 6 1993 99
## 7 1994 35
## 8 1995 89
## 9 1997 82
## 10 1998 63
## # … with 11 more rowsWalk Through Notes:
- First we filter the data for flows above 1000 cfs and dated from March to July (most of the monsoons occur from august to October).
- Then we simply count the number of dates that are left by year.
We now have the variable we want to predict. Now we need predictors.
Getting the predictive variables.
There are many variables that affect reservoir volume. Current Reservoir volume and snowpack are probably two of the more important variables that are reliably measured. Snowpack tells us how much water will eventually be coming into the reservoir in the spring and reservoir volume tells us how much water is currently in the reservoir and also how much water we will need to cause a spill. Spring rain would be another variable that would be great to include, but I don’t want to rely on weather forecasts that far out. So we will stick with snowpack and current reservoir volume.
Snow Depth
The SNOTEL (Snow Telemetry) network provides snow data for 730 sites across the country. There are several in the Dolores watershed that can give us a bunch of data on snowpack. For this we will use the handy dandy package library(RNRCS) (the NRCS is the government agency in the Department of Agriculture that manages the SNOTEL network)
Finding All Snotel Sites in the Dolores Watershed
First we need to find which SNOTEL sites are within the Dolores River watershed to inform how much water could end up in McPhee reservoir.
library(RNRCS)
meta_data<-grabNRCS.meta(ntwrks = c("SNTL", "SNTLT", "SCAN"))
meta_data_unlist<-meta_data[[1]]%>%
as_tibble()
meta_data_unlist## # A tibble: 876 x 13
## ntwk wyear state site_name ts start enddate latitude longitude
## <chr> <fct> <fct> <chr> <fct> <fct> <fct> <fct> <fct>
## 1 SNTL 2019 AK Frostbit… "" 2019… 2100-J… " 61.7… -149.27
## 2 SNTL 2019 AK McGrath "" 2019… 2100-J… " 62.9… -155.61
## 3 SNTL 2019 UT Parleys … "" 2019… 2100-J… " 40.7… -111.61
## 4 SNTL 2018 AK East Pal… "" 2018… 2100-J… " 61.6… -149.10
## 5 SNTL 2018 AK Galena AK "" 2018… 2100-J… " 64.7… -156.71
## 6 SNTL 2018 MT JL Meadow "" 2017… 2100-J… " 44.7… -113.12
## 7 SNTL 2018 NV ONeil Cr… "" 2018… 2100-J… " 41.8… -115.08
## 8 SNTL 2017 AK Flower M… "" 2017… 2100-J… " 59.4… -136.28
## 9 SNTL 2017 CA Fredonye… "" 2016… 2100-J… " 40.6… -120.61
## 10 SNTL 2017 UT Bobs Hol… "" 2016… 2100-J… " 38.9… -112.15
## # … with 866 more rows, and 4 more variables: elev_ft <fct>, county <fct>,
## # huc <fct>, site_id <chr>grabNRCS.meta - grabs all sites within the Snotel (“SNTL”), Snotel Lite, (“SNTLT”), and Scan (“SCAN”) systems. I’m really just interested in the snotel sites, but I wasn’t really sure what Snotel Lite or Scan was so I loaded them just in case. grabNRCS returns a single list, se we subset just the first item of the list.
dolores_sites<-meta_data_unlist%>%
filter(state =="CO", str_detect(huc, "140300"))%>%
mutate(site_id_num = as.numeric(str_match_all(site_id, "[0-9]+")))
dolores_sites## # A tibble: 6 x 14
## ntwk wyear state site_name ts start enddate latitude longitude
## <chr> <fct> <fct> <chr> <fct> <fct> <fct> <fct> <fct>
## 1 SNTL 2011 CO Black Me… "" 2011… 2100-J… " 37.7… -108.18
## 2 SNTL 2005 CO Sharksto… "" 2004… 2100-J… " 37.5… -108.11
## 3 SNTL 1986 CO El Dient… "" 1985… 2100-J… " 37.7… -108.02
## 4 SNTL 1986 CO Scotch C… "" 1985… 2100-J… " 37.6… -108.01
## 5 SNTL 1980 CO Lizard H… "" 1979… 2100-J… " 37.8… -107.92
## 6 SNTL 1980 CO Lone Cone "" 1979… 2100-J… " 37.8… -108.20
## # … with 5 more variables: elev_ft <fct>, county <fct>, huc <fct>,
## # site_id <chr>, site_id_num <dbl>dolores_sites - We then filter all sites in Colorado (“CO”) and detect all of the huc values with 140300. To pull data from the sites we will need a site ID number. The Meta data has the site IDs in a string that contains characters and numbers.
dolores_site_ids<-dolores_sites%>%
pull(site_id_num)%>%
unlist()%>%
as.numeric()
dolores_site_ids## [1] 1185 1060 465 739 586 589dolores_site_ids - The last step is to convert the site_id_num variable into a vector so that we can pull data from each site in the next step.
Pulling the data from the snotel sites
Function to Pull All Sites
First we need a function that we can loop over each of the snotel site IDs and pull data for each of one. grabNRCS.data does not allow you to pull data from more than one site at a time.
get_snotl_data<-function(site_id){
grabNRCS.data(network = "SNTL",
site_id = site_id,
timescale = "daily",
DayBgn = '1985-01-01',
DayEnd = Sys.Date()
)%>%
as_tibble()%>%
mutate(site_id_num = site_id)
}To get data from each site we need to use grabNRCS.data() function. grabNRCS.data takes:
network- the network we want to pull from.- the
site_idwhich we produced a list of in the last step. DayBgn- The first day you want to pull data from.DayEnd- the Last date you want data from. Here we use the system data (Today).
We then convert the data to a tibble for better output in Rstudio and add a column with mutate with the site_id number for working with the data later.
Getting ALL the data with lapply
all_sntl_data<-lapply(dolores_site_ids, get_snotl_data)%>%
bind_rows()
all_sntl_data## # A tibble: 58,531 x 22
## Date Air.Temperature… Air.Temperature… Air.Temperature…
## <chr> <int> <int> <int>
## 1 2012… 46 52 42
## 2 2012… 46 53 42
## 3 2012… 52 62 44
## 4 2012… 52 61 46
## 5 2012… 52 64 46
## 6 2012… 54 64 47
## 7 2012… 53 64 49
## 8 2012… 52 61 47
## 9 2012… 54 63 49
## 10 2012… 50 61 42
## # … with 58,521 more rows, and 18 more variables:
## # Air.Temperature.Observed..degF..Start.of.Day.Values <int>,
## # Precipitation.Accumulation..in..Start.of.Day.Values <dbl>,
## # Snow.Depth..in..Start.of.Day.Values <int>,
## # Snow.Water.Equivalent..in..Start.of.Day.Values <dbl>,
## # Soil.Moisture.Percent..4in..pct..Start.of.Day.Values <dbl>,
## # Soil.Moisture.Percent..8in..pct..Start.of.Day.Values <dbl>,
## # Soil.Moisture.Percent..20in..pct..Start.of.Day.Values <dbl>,
## # Soil.Moisture.Percent..40in..pct..Start.of.Day.Values <dbl>,
## # Soil.Temperature.Observed..4in..degF..Start.of.Day.Values <int>,
## # Soil.Temperature.Observed..8in..degF..Start.of.Day.Values <int>,
## # Soil.Temperature.Observed..20in..degF..Start.of.Day.Values <int>,
## # Soil.Temperature.Observed..40in..degF..Start.of.Day.Values <int>,
## # site_id_num <dbl>,
## # Soil.Moisture.Percent..2in..pct..Start.of.Day.Values <dbl>,
## # Soil.Temperature.Observed..2in..degF..Start.of.Day.Values <int>,
## # Wind.Direction.Average..degree. <int>, Wind.Speed.Average..mph. <dbl>,
## # Wind.Speed.Maximum..mph. <dbl>lapply takes:
dolores_site_idsa vector of values to loop over.get_snotl_dataa function to inject each of the vector values into one at a time.
lapply returns a list so the final step is to bind_rows which merges all of the columns.
Cleaning the Data
I’m going to predict number of days the Dolores Runs in a year, so I need to convert the snow data from daily data from multiple sites to annual data from all sites. I’m assuming that max snow water equivalant correlates best with runoff in this dataset so we will use that to summarize each year.
se_site<-all_sntl_data%>%
select(Date, Snow.Depth..in..Start.of.Day.Values, Snow.Water.Equivalent..in..Start.of.Day.Values, site_id_num)%>%
mutate(
date = as.Date(Date)
)%>%
rename(snow_depth = 2, snow_water_eq=3)%>%
filter(site_id_num %in% c(465, 586, 589, 739))
se_site## # A tibble: 50,182 x 5
## Date snow_depth snow_water_eq site_id_num date
## <chr> <int> <dbl> <dbl> <date>
## 1 1986-08-05 NA NA 465 1986-08-05
## 2 1986-08-06 NA 0 465 1986-08-06
## 3 1986-08-07 NA 0 465 1986-08-07
## 4 1986-08-08 NA 0 465 1986-08-08
## 5 1986-08-09 NA 0 465 1986-08-09
## 6 1986-08-10 NA 0 465 1986-08-10
## 7 1986-08-11 NA 0 465 1986-08-11
## 8 1986-08-12 NA 0 465 1986-08-12
## 9 1986-08-13 NA 0 465 1986-08-13
## 10 1986-08-14 NA 0 465 1986-08-14
## # … with 50,172 more rows- First we select the columns that contain the Date, Snow Depth and Snow water Equivalant. I don’t end up using Snow depth because it only goes back to around 2000 for all sites while snow water equivalant goes back to 1986.
mutateadd a date field that is formatted as a date.renamesnow depth and snow water equivalant to something more manageable.- The last thing we do is subset the sites to only those that have been around since 1986 using
filter.
Let’s take a look at these sites really quick to see how they look.
se_site%>%
filter(year(date)>2007)%>%
ggplot(aes(date, snow_water_eq, color = as.factor(site_id_num)))+
geom_line(size = 1.5, alpha = 0.5)+
theme_light()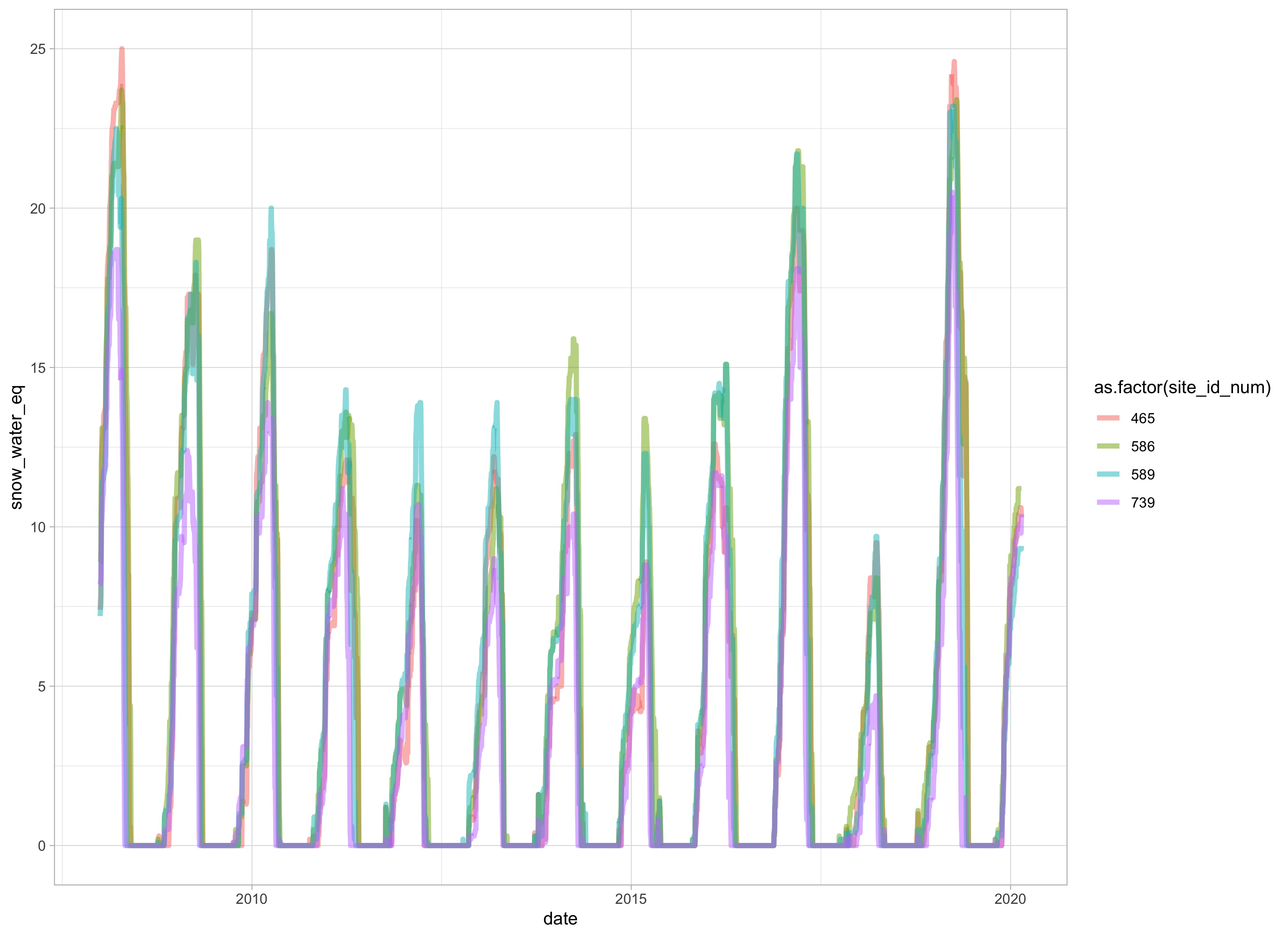
se_site_year<-se_site%>%
group_by(year(date), site_id_num)%>%
summarize(max_se = max(snow_water_eq, na.rm = T))%>%
ungroup()%>%
rename(year=1)
se_site_year## # A tibble: 142 x 3
## year site_id_num max_se
## <dbl> <dbl> <dbl>
## 1 1985 586 21.6
## 2 1985 589 23.5
## 3 1986 465 6.3
## 4 1986 586 19.8
## 5 1986 589 20.8
## 6 1986 739 5.1
## 7 1987 465 19.1
## 8 1987 586 19.1
## 9 1987 589 22
## 10 1987 739 20.1
## # … with 132 more rows- The next several steps are where the sausage is made. First I want the max value for snow water equivalant for each site per year. To do this, we
group_byyear(date)which uses thedatefield to group the data and then. We also want to group by site_id_num so we add that to thegroup_byas well. - We can then summarize by our groups. Here we want to make a variable caleed max_se which is the max of the
snow_water_eqfield within the group we described above. - Lastly we
ungroupthe data (group_by can cause nasty problems if not ungrouped) and rename theyear(date)column toyear.
Combining a subset of sites
To simplify things we will average the sites. I think if I did this again I would not average the sites but have each site be a predictor.
avg_snwater_eq<-se_site_year%>%
filter(year>1986)%>%
group_by(year)%>%
summarize(avg_snow_water_e = mean(max_se))%>%
ungroup()
avg_snwater_eq## # A tibble: 34 x 2
## year avg_snow_water_e
## <dbl> <dbl>
## 1 1987 20.1
## 2 1988 11.4
## 3 1989 16.3
## 4 1990 9.28
## 5 1991 16.5
## 6 1992 15.5
## 7 1993 25.4
## 8 1994 12.3
## 9 1995 21.8
## 10 1996 13.6
## # … with 24 more rows- Only 4 of the 6 sites in the basin have values all the way back to 1986, the year that the McPhee Dam was put in.
- We then
group_byyear because we want one value per year. - Then
summarizethe max snow water equivalent (max_se) - created in the last step - as a mean of the four site maxes per year. - And then, as always,
ungroup.
Summary of Snotel Snow depth
Now we have our first predictive variable: average max snow water equivalent four three snotel sites in the Dolores River watershed.
The steps we took to get that data: 1. We used the RNRCS package to… 2. Get all sites in the Snotel Network with grabNRCS.meta() 3. We then subset those sites to just those in the Dolores watershed. 4. We then used the site_ids to get daily snow data from each site. 5. We combined each sites data into one dataframe. 6. We then grouped the data to get the max snow water equivalent for each site for each year. 7. We then averaged the maxes from four sites to get one average snow water equivalent per year.
Prediction of Future Snowpack
We’ll use the {fable} package for timeseries forecasting.
library(fable) #For predictionFor forecasting we need an average of all four sites per day. We want to predict what the average will be.
avg_sn<-se_site%>%
filter(year(Date)>1987)%>%
group_by(Date)%>%
summarize(avg_sn_eq = mean(snow_water_eq, rm.na = T))%>%
ungroup()%>%
mutate(Date = as.Date(Date))
avg_sn## # A tibble: 11,741 x 2
## Date avg_sn_eq
## <date> <dbl>
## 1 1988-01-01 4.1
## 2 1988-01-02 4.18
## 3 1988-01-03 4.38
## 4 1988-01-04 4.53
## 5 1988-01-05 4.82
## 6 1988-01-06 5.4
## 7 1988-01-07 5.6
## 8 1988-01-08 5.65
## 9 1988-01-09 5.8
## 10 1988-01-10 5.9
## # … with 11,731 more rows- We filter years after 1987 because we don’t want the year that they were filling the reservior to influence the prediction.
- Next we group by
Datewhich is daily and thensummarize, averaging all four sites for that day. Weungroupto prevent weird stuff from happening. - And lastly we convert the
Datecolumn, which for some reaon was converted to acharacter, to an actual date withas.Date.
Let see what it looks like:
avg_sn%>%
ggplot(aes(Date, avg_sn_eq))+
geom_line(color = "#1AB8FF", size = 2, alpha = 0.5)+
labs(title = "Average Snow Water Equivalant",
subtitle = "From four Snotel Sites in the Dolores River watershed",
y = "Avg. Snow Water Equivalant")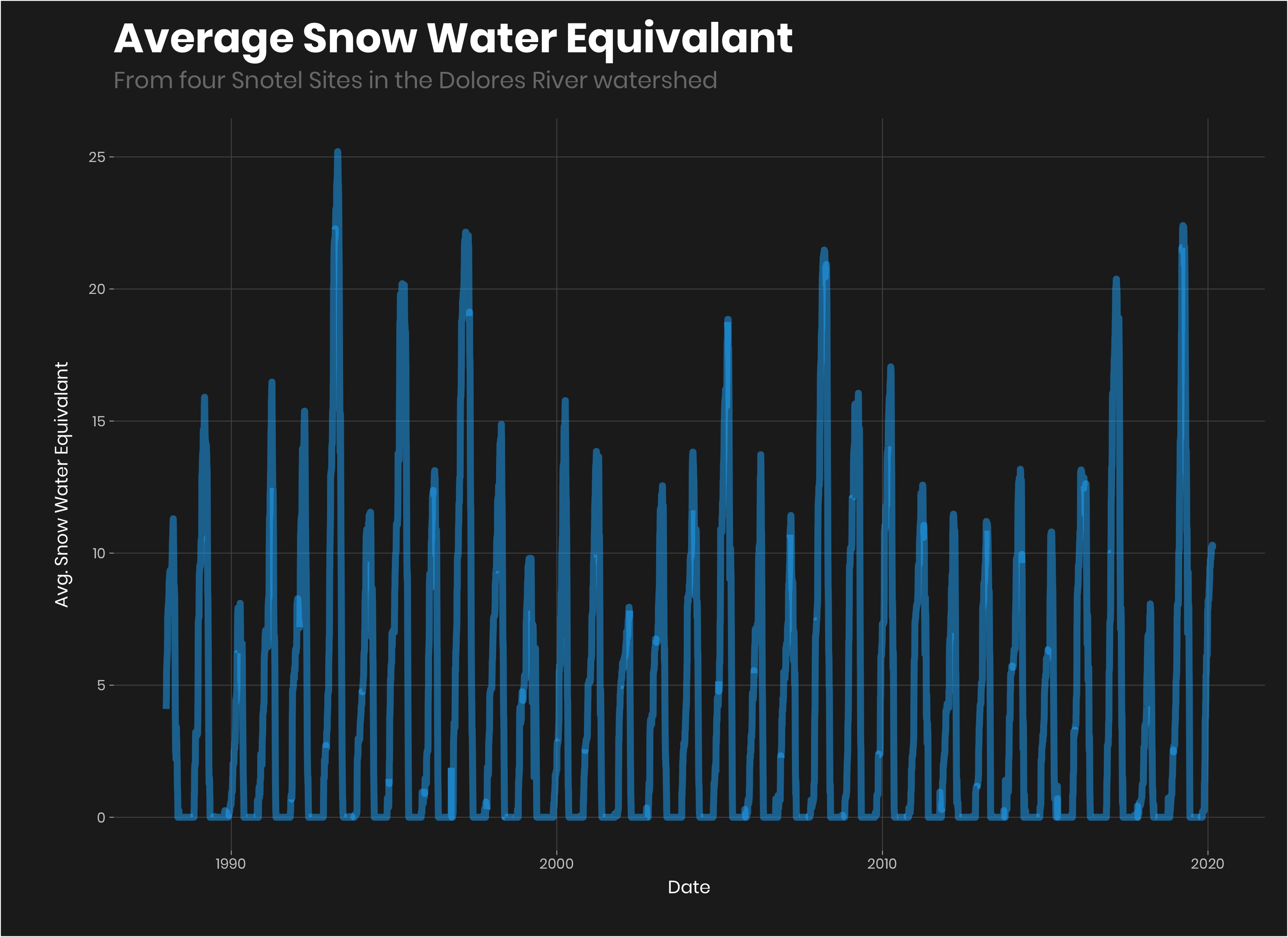
To do a timeseries forcast we need to convert the data into tsibble format. I don’t know a ton about tsibbles. I think they are basically just a tibble with an time index.
avg_sn_ts<-avg_sn%>%
as_tsibble(index = Date)Next we will make our model. I by no means am an expert at forecasting (this took me an embarassingly long time). Many of my initial tries failed using a variety of other models. I finally got an auto ARIMA forecast to work using fourier. If you want to learn more about forecasting, check out Rob Hyndman’s Blog.
fit<-avg_sn_ts%>%
model(
arima = ARIMA(avg_sn_eq~fourier("year", K = 15))
)
fit%>%
forecast(h = "3 months")%>%
autoplot(avg_sn_ts%>%filter(year(Date)>2018))+
geom_line(color = "#ffffff", size = 2)+
labs(title = "Snowpack Prediction",
subtitle = "Dolores River watershed Snotel sites",
y = "Avg. Snow Whater Equivalant")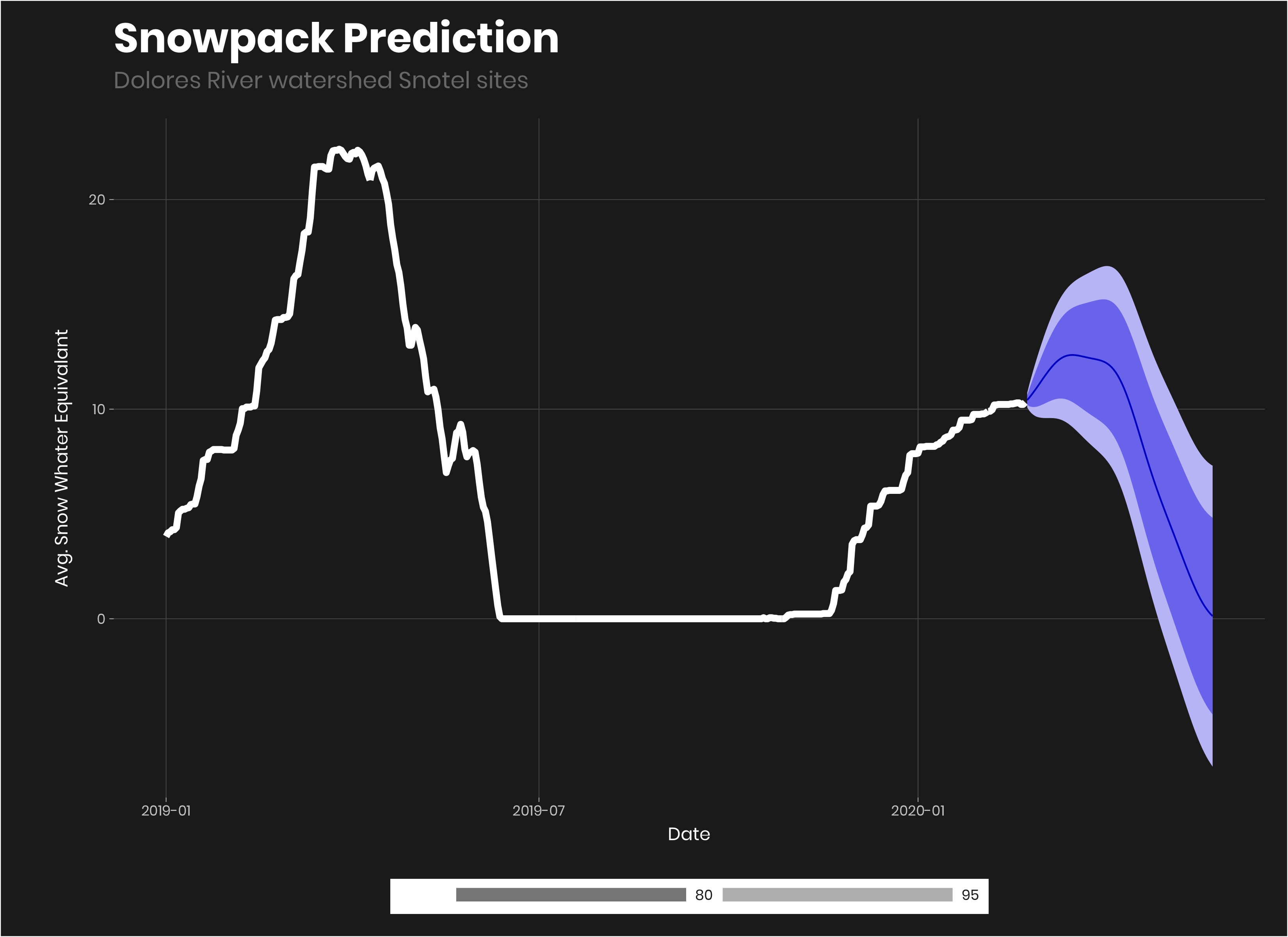
Looking at the forecast in the autoplot() which is just an easy ggplot for forecasting, it looks to me that the forecast looks pretty good, but might be a bit optimistic for the average. But it’s hard to tell. We could get a lot of snow in the end of February or March. It gives us a good range to work with and use in our predictions.
To use in our prediction we need a max, min and average value for the max snowpack.
pred_sn_pk<-fit%>%
forecast(h = "6 months")%>%
filter(avg_sn_eq == max(avg_sn_eq))%>%
mutate(ci_90 = hilo(.distribution, 90))%>%
mutate(pred_lower = ci_90$.lower, pred_upper = ci_90$.upper)%>%
as_tibble()%>%
rename(pred_avg_sn_eq = avg_sn_eq)%>%
select(Date, pred_avg_sn_eq, pred_lower, pred_upper)%>%
pivot_longer(-Date, names_to = "estimated_eq", values_to = "avg_snow_water_e")%>%
mutate(year = year(Date))%>%
select(-Date)
pred_sn_pk## # A tibble: 3 x 3
## estimated_eq avg_snow_water_e year
## <chr> <dbl> <dbl>
## 1 pred_avg_sn_eq 12.6 2020
## 2 pred_lower 9.70 2020
## 3 pred_upper 15.5 2020- Use our model to
forecast()six months out. filterout themaxvalue for the average (this isn’t perfect but it will work for our purposes).- Add a column with higher and lower values based on a 90% confidence interval using the
hilofunction. - convert to a tibble.
- Select only the values we want.
pivot_longerso that we have ayearcolumn and a snow_water_equivalant column.- Make a
yearcolumn. - drop the
Datecolumn.
Reservoir Volume
The next variable we need is reservoir volume. ### Get Daily McPhee Reservoir Volumes
bor_data<-grabBOR.data(site_id = "MPHC2000",
timescale = 'daily',
DayBgn = '1985-01-01',
DayEnd = Sys.Date())%>%
as_tibble()%>%
mutate(date = as.Date(Date),
res_volume = as.numeric(`Reservoir Storage Volume (ac_ft) Start of Day Values`))%>%
select(date, res_volume)
bor_data## # A tibble: 12,826 x 2
## date res_volume
## <date> <dbl>
## 1 1985-01-01 44997
## 2 1985-01-02 45079
## 3 1985-01-03 45161
## 4 1985-01-04 45189
## 5 1985-01-05 45207
## 6 1985-01-06 45262
## 7 1985-01-07 45372
## 8 1985-01-08 45463
## 9 1985-01-09 45601
## 10 1985-01-10 45721
## # … with 12,816 more rowsWe use the {RNRCS} package again to get Bureau of Reclemation (BOR - the agency that managed the McPhee dam)data.
- The
{RNRCS}package has a function to grab Bureau of Reclemation datagrabBOR.data(). It works much the same way as thegrapNRCS.data()function that we wrapped our own function around above. However this time, we only needed one site so I just looked it up and put it in instead of using a meta function to get sites like we did with the snotel data. - We convert the data to a tibble (df) convert the date to date format and convert the volume into numeric.
- We then subset the columns to just date and reservoir volume.
Summarize data to yearly
res_vol<-bor_data%>%
filter(month(date) %in% c(01, 02))%>%
group_by(year(date))%>%
summarize(min_vol = min(res_volume, na.rm = T))%>%
ungroup()%>%
rename(year = 1)%>%
filter(year>1986)
res_vol## # A tibble: 34 x 2
## year min_vol
## <dbl> <dbl>
## 1 1987 293309
## 2 1988 324234
## 3 1989 319239
## 4 1990 274597
## 5 1991 241536
## 6 1992 297471
## 7 1993 304632
## 8 1994 310609
## 9 1995 268308
## 10 1996 312568
## # … with 24 more rowsHere we subset the data to just the winter months and then summarize by year by getting the minimum value for all three months (I originally averaged the months, but getting the min gives a better prediction).
- First we filter January and February to get that years lowest reservoir values prior to runoff.
- We then
group_by()year and then get the minimul volume from each of the two months. - then we rename the first column to year instead of
year(date)and filter out all years before 1987.
Building the Model
Combining the variables and the datasets.
var_df<-predicted_variable%>%
full_join(avg_snwater_eq)%>%
full_join(res_vol)%>%
arrange(year)%>%
mutate(raftable_releases = ifelse(is.na(raftable_releases), 0, raftable_releases))
var_df_train<-var_df%>%
filter(year!=2020)
var_df_test<-var_df%>%
filter(year==2020)
var_df_train## # A tibble: 33 x 4
## year raftable_releases avg_snow_water_e min_vol
## <dbl> <dbl> <dbl> <dbl>
## 1 1987 87 20.1 293309
## 2 1988 13 11.4 324234
## 3 1989 17 16.3 319239
## 4 1990 0 9.28 274597
## 5 1991 9 16.5 241536
## 6 1992 61 15.5 297471
## 7 1993 99 25.4 304632
## 8 1994 35 12.3 310609
## 9 1995 89 21.8 268308
## 10 1996 0 13.6 312568
## # … with 23 more rows- To join the datasets we use a full join because not every year has a raftable release and therefore has some year missing. Both of the other datasets have data for each year and a full join fills any missing variables with NA but keeps all observations.
- The years with NA values for raftable release should be 0, because those are the years without a raftable release and we want to th model to predict when those years are as well.
- Lastly we make two datasets one to train the model, which includes all years except this year 2020, and test model this year.
Note: typically you want to split the data into training and testing datasets so you can train your model with the training dataset and then evaluate it with the testing dataset that it hasn’t seen. But here we don’t really care that much because this is just fun and we can predict this upcoming release and see if it is right.
Random Forest Using caret
library(caret)
set.seed(1234)
control <- trainControl(method="cv", number=10)
rf_model <- train(raftable_releases~avg_snow_water_e+min_vol,
data=var_df_train,
method="rf",
tuneLength=3,
trControl=control)## note: only 1 unique complexity parameters in default grid. Truncating the grid to 1 .rf_model## Random Forest
##
## 33 samples
## 2 predictor
##
## No pre-processing
## Resampling: Cross-Validated (10 fold)
## Summary of sample sizes: 29, 31, 29, 29, 29, 30, ...
## Resampling results:
##
## RMSE Rsquared MAE
## 15.69357 0.6885501 13.05262
##
## Tuning parameter 'mtry' was held constant at a value of 2Here we build a cross validated random forest regression model to go predict number of raftable days.
meothod = "repeatedcv"is repeated cross validation where we do 10 cross validations 3 times with a random search.- We use the
train()functio from caret to fit the model using both predictive variablesavg_snow_water_eandmin_vol. - We use Random Forest algorythm
"rf"with 2000 trees. - we try 3 mtry depths (thgere are only two because we have only two variables, but tuneLength of two was not tuning at all for some reason).
Evaluating the model
WARNING: this is not how you should evaluate a model typically like I said above.
var_df_train%>%
mutate(prediction = stats::predict(rf_model, .))%>%
ggplot(aes(raftable_releases, prediction))+
geom_point(color = "#1AB8FF", size =4, alpha = 0.6)+
geom_text(aes(label=year),hjust=-0.3, vjust=-0.3, color = "#FFFFFF")+
labs(title = "Random Forest: Predicted vs Actual Raftable Release Days",
x = "Actual Release Days",
y = "Predicted Release Days",
subtitle = "Sudo-testing accuracy of RF model")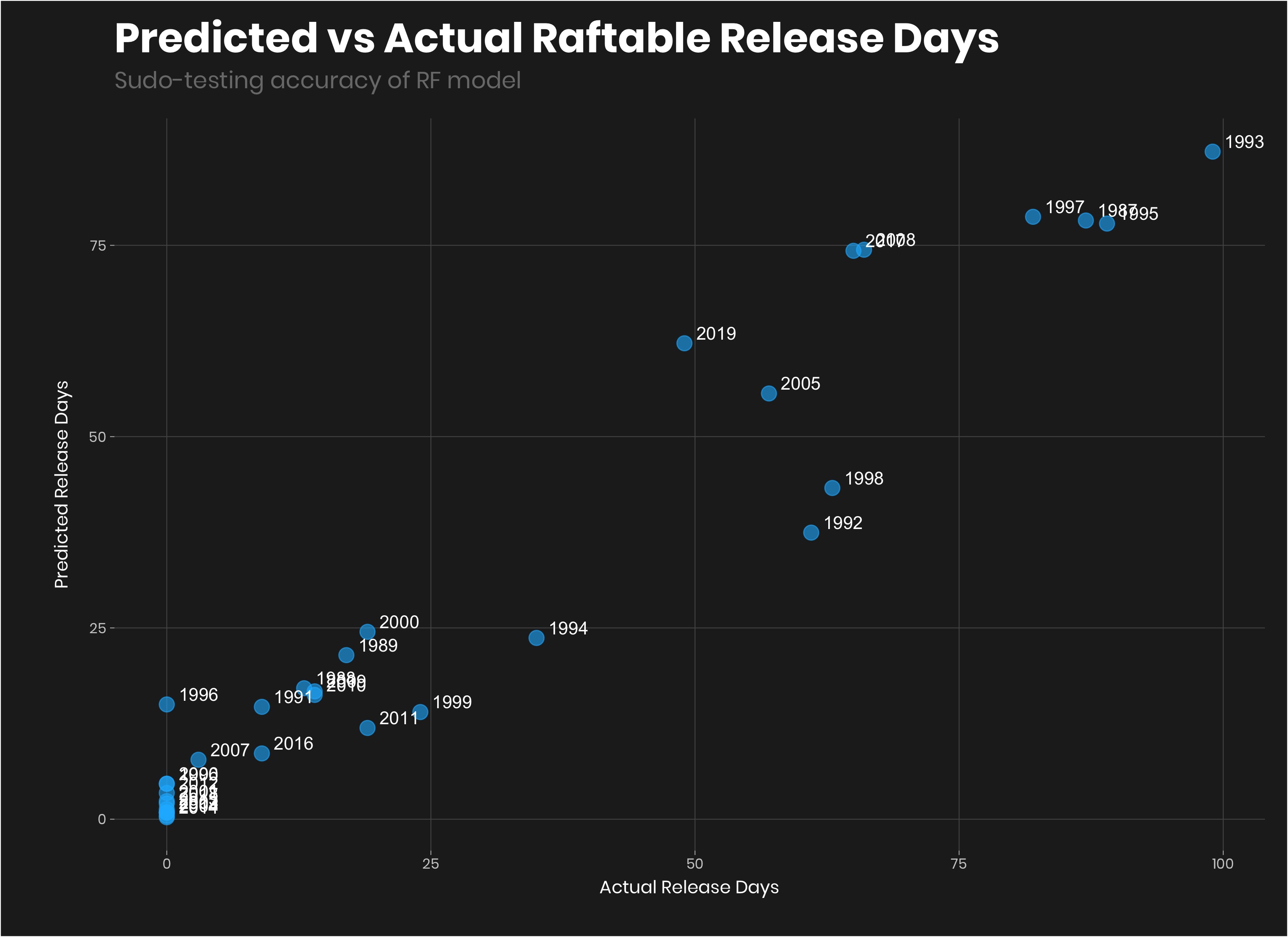
Pretty good. Again, I keep saying this becuase it is really important, we would want to make sure that the model works in out of training sample, but with this dataset, we don’t have enough data to do that.
So what about this year
var_df_test%>%
mutate(prediction = predict(rf_model, .))## # A tibble: 1 x 5
## year raftable_releases avg_snow_water_e min_vol prediction
## <dbl> <dbl> <dbl> <dbl> <dbl>
## 1 2020 0 10.4 287807 4.71So far the model predicts we will have 4.7078333 days of raftable flows. I’d say that is within the margin of error of 0 days looking at past predictions. But what if we get more snow? Given that it is February and Peak snow is typically around mid March would be a good assumption.
Let’s add the snotel forcast from the ARIMA model above to the predicted dataset.
pred_sn_pk_1<-pred_sn_pk%>%
mutate(min_vol = var_df%>%
filter(year == 2020)%>%
pull(min_vol))
test_data<-var_df%>%
filter(year==2020)%>%
select(-raftable_releases)%>%
mutate(estimated_eq = "current")%>%
bind_rows(pred_sn_pk_1)
test_data## # A tibble: 4 x 4
## year avg_snow_water_e min_vol estimated_eq
## <dbl> <dbl> <dbl> <chr>
## 1 2020 10.4 287807 current
## 2 2020 12.6 287807 pred_avg_sn_eq
## 3 2020 9.70 287807 pred_lower
## 4 2020 15.5 287807 pred_upperNow let’s use the prediction for current, pred_avg_sn_eq, pred_lower, and pred_upper
test_data%>%
mutate(prediction = stats::predict(rf_model, .))## # A tibble: 4 x 5
## year avg_snow_water_e min_vol estimated_eq prediction
## <dbl> <dbl> <dbl> <chr> <dbl>
## 1 2020 10.4 287807 current 4.71
## 2 2020 12.6 287807 pred_avg_sn_eq 6.60
## 3 2020 9.70 287807 pred_lower 3.09
## 4 2020 15.5 287807 pred_upper 23.0That looks pretty good.
- Current Prediction is 4.71 days.
- Predicted is 6.6 at the average forecast snowdepth.
- 3.09 is the low end. And 23 days if we get dumped on from now until April 1.
Let’s try something a bit simpler, like a linear model.
model_lm<-glm(raftable_releases~avg_snow_water_e+min_vol, family = "poisson", data = var_df_train)
##saveRDS(model_lm, "output/models/lm_model.rds")
summary(model_lm)##
## Call:
## glm(formula = raftable_releases ~ avg_snow_water_e + min_vol,
## family = "poisson", data = var_df_train)
##
## Deviance Residuals:
## Min 1Q Median 3Q Max
## -6.033 -3.211 -2.308 1.997 6.488
##
## Coefficients:
## Estimate Std. Error z value Pr(>|z|)
## (Intercept) -2.0553926585 0.2693758160 -7.63 0.0000000000000234
## avg_snow_water_e 0.1900049675 0.0072692190 26.14 < 0.0000000000000002
## min_vol 0.0000076058 0.0000008358 9.10 < 0.0000000000000002
##
## (Intercept) ***
## avg_snow_water_e ***
## min_vol ***
## ---
## Signif. codes: 0 '***' 0.001 '**' 0.01 '*' 0.05 '.' 0.1 ' ' 1
##
## (Dispersion parameter for poisson family taken to be 1)
##
## Null deviance: 1292.88 on 32 degrees of freedom
## Residual deviance: 446.98 on 30 degrees of freedom
## AIC: 563.07
##
## Number of Fisher Scoring iterations: 6This model looks pretty good.
model_lm%>%
augment(data = var_df_train, type.predict = "response")%>%
ggplot(aes(raftable_releases, .fitted))+
geom_point(color = "#1AB8FF", size =4, alpha = 0.6)+
geom_text(aes(label=year),hjust=-0.3, vjust=-0.3, color = "#FFFFFF")+
labs(title = "Linear Model: Predicted vs Actual Raftable Release Days",
subtitle = "Actual Values vs. Predicted Values",
y = "Predicted Values",
x = "Actual Raftable Release Days")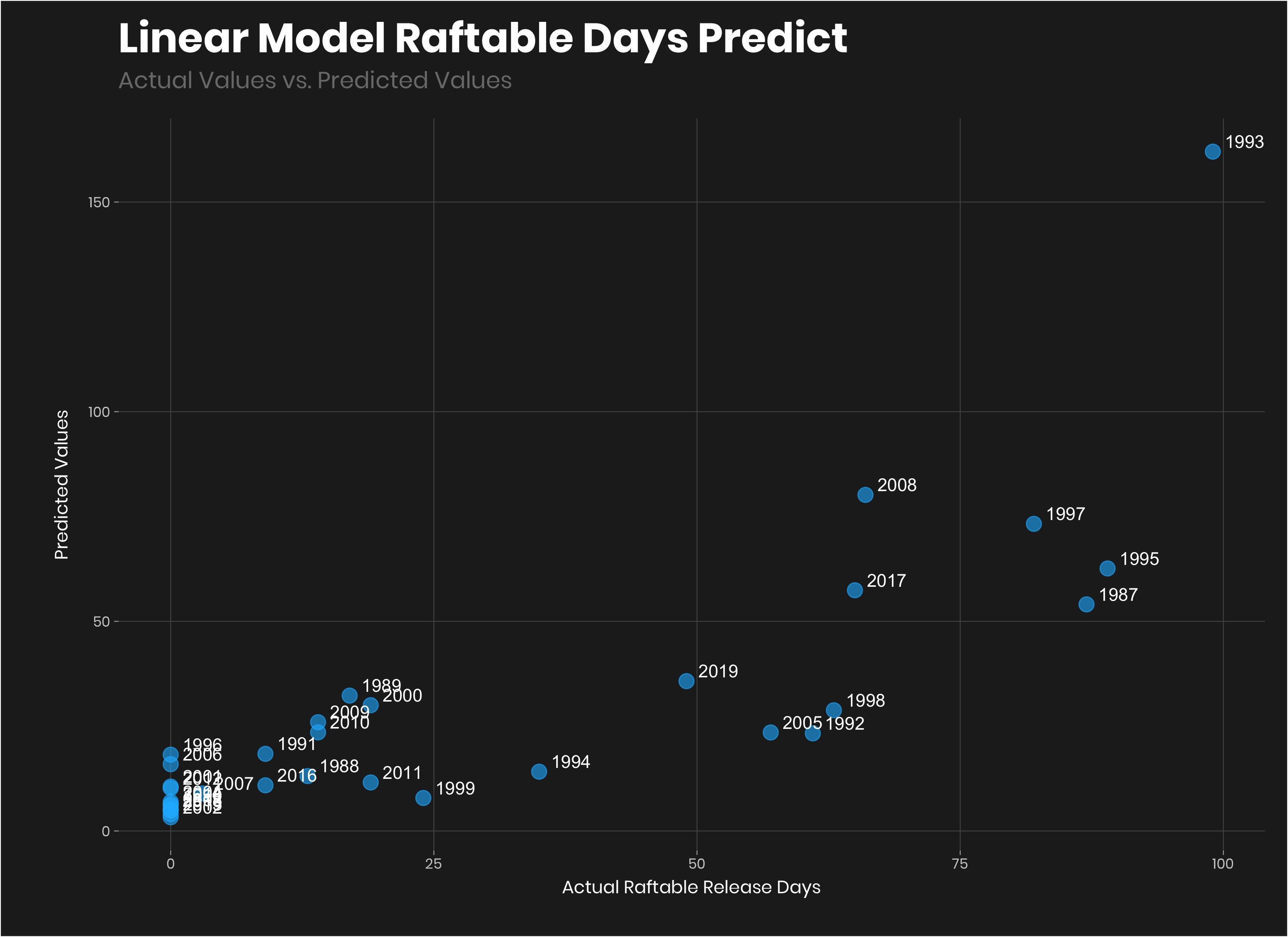
model_lm%>%
augment(newdata = test_data, type.predict = "response")## # A tibble: 4 x 6
## year avg_snow_water_e min_vol estimated_eq .fitted .se.fit
## <dbl> <dbl> <dbl> <chr> <dbl> <dbl>
## 1 2020 10.4 287807 current 8.25 0.620
## 2 2020 12.6 287807 pred_avg_sn_eq 12.5 0.767
## 3 2020 9.70 287807 pred_lower 7.21 0.575
## 4 2020 15.5 287807 pred_upper 21.6 0.986That looks more rational than the random forest model. The results:
- Current snowpack = 8.25 days
- predicted peak snowpack = 12.5 days.
- predicted lower snowpack = 7.21 (this really isn’t rational considering current is already greater than it).
- predicted upper snowpack = 21.6 days (let it snow let it snow).
I’ll continue to run this as the winter progresses. We now have a model that we can push into production to predict future data sets.